Notes for Class 12 Biology Chapter 11 Biotechnology Principles and Processes
Commerce students can refer to the Biotechnology Principles and Processes Notes Class 12 Biology given below which is an important chapter in class 12 Biology book. These notes and important questions and answers have been prepared based on the latest CBSE and NCERT syllabus and books issued for the current academic year. Our team of Biology teachers have prepared these notes for the benefit of students so that you can read these revision notes and understand each topic carefully.
Biotechnology Principles and Processes Notes Class 12 Biology
Refer to the notes and important questions given below for Biotechnology Principles and Processes which are really useful and have been recommended by Class 12 Biology teachers. Understanding the concepts in detail and then solving questions by yourself will help you to learn all topics given in your NCERT Books for Class 12 Biology.
• Biotechnology is the technique of using live organisms or their enzymes for products & processes useful to humans.
• The European Federation of Biotechnology (EFB) defines Biotechnology as ‘the integration of natural science and organisms, cells, parts thereof, and molecular analogues for products and services’.
Biotechnology deals with:
– Microbe-mediated processes (making curd, bread, wine etc).
– In vitro fertilization (test-tube baby programme).
– Synthesis and using of a gene.
– Preparation of DNA vaccine.
– Correcting a defective gene.
PRINCIPLES OF BIOTECHNOLOGY
Core techniques of modern biotechnology
• Genetic engineering : The technique in which genetic material (DNA & RNA) is chemically altered and
introduced into host organisms to change the phenotype.
• Bioprocess engineering : Maintenance of sterile ambience in chemical engineering processes for growing desired microbe/eukaryotic cell for the manufacture of antibiotics, vaccines, enzymes etc.
Basic steps in genetically modifying an organism
a) Identification of DNA with desirable genes : Traditional hybridisation leads to inclusion and multiplication of undesirable genes along with desired genes. In genetic engineering, only desirable genes are introduced.
b) Introduction of the identified DNA into the host : A vector DNA such as plasmid is used to deliver an alien piece of DNA into the host organism.
c) Maintenance of introduced DNA in the host and transfer of the DNA to its progeny : A piece of alien DNA has no the sequence called Origin of replication (ori) needed for starting replication. So, it cannot multiply itself in the progeny cells of the organism. Hence alien DNA is integrated into the recipient genome (it has ori).
It multiplies & inherits along with host DNA.
• The process of joining and inserting a foreign piece of DNA into a host organism to produce new genetic combinations is called recombinant DNA technology.
• First recombinant DNA (rDNA) was produced by Stanley Cohen & Herbert Boyer (1972).
• They isolated an antibiotic resistance gene (piece of DNA) from a plasmid of Salmonella typhimurium. It was linked with a plasmid vector and transferred into E. coli.
As a result, the gene was expressed & multiplied in E. coli.
TOOLS OF RECOMBINANT DNA TECHNOLOGY
1. Restriction Enzymes (‘molecular scissors’)
– The enzymes that cut DNA at specific sites into fragments.
– They belong to a class of enzymes called nucleases.
– In 1963, two enzymes responsible for restricting growth of bacteriophage in E. coli were isolated. One enzyme added methyl groups to DNA. The other (restriction endonuclease) cut DNA.
– More than 900 restriction enzymes have been isolated from over 230 strains of bacteria.
Naming of the restriction enzymes :
– First letter indicates genus. The second two letters indicate species of prokaryotic cell from which they were isolated.
E.g. EcoRI comes from E. coli RY 13 (R = the strain.
Roman numbers = the order in which the enzymes were isolated from that strain of bacteria).
Types of Restriction enzymes :
• Exonucleases : They remove nucleotides from the ends of the DNA.
• Endonucleases :
– They cut at specific positions within the DNA. E.g. EcoRI.
– They bind to specific recognition sequence of the DNA and cut the two strands at specific points.
– The first restriction endonuclease is Hind II. It cuts DNA molecules by recognizing a specific sequence of 6 base pairs. This is called the recognition sequence for Hind II.
– Restriction endonuclease recognizes a specific palindromic nucleotide sequences in the DNA. It is a sequence of base pairs that read the same on the two strands in 5′ → 3′ direction and in 3′ → 5′ direction. E.g.
Palindromic nucleotide sequence for EcoRI is
5′ —— GAATTC —— 3′
3′ —— CTTAAG —— 5′
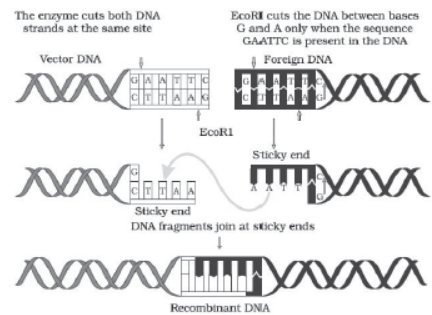
– Restriction enzymes cut the strand a little away from the centre of the palindrome sites, but between the same two bases on the opposite strands. This leaves single stranded
overhanging stretches at the ends. They are called sticky ends. They form H-bonds with their complementary cut counterparts. This stickiness facilitates action of the enzyme DNA ligase.
– When cut by the same restriction enzyme, the resultant DNA fragments have the same kind of sticky-ends and these are joined together by DNA ligases.
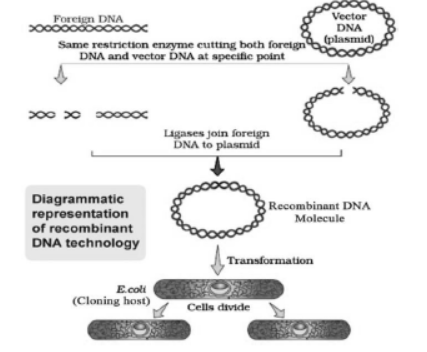
2. Cloning Vector
It is a DNA molecule that can carry a foreign DNA segment and replicate inside the host cells.
E.g. Plasmids, bacteriophages etc.
– Plasmids are autonomously replicating circular extrachromosomal DNA of bacteria. Some plasmids have only
1-2 copies per cell. Others have 15-100 copies per cell.
– Bacteriophages (high number per cell) have very high copy numbers of their genome within the bacterial cells.
– When the cloning vectors are multiplied in the host, the linked piece of DNA is also multiplied to the numbers equal to the copy number of the vectors.
Features required for cloning into a vector
a. Origin of replication (ori)
– This is a sequence where replication starts.
– A piece of DNA linked to ori can replicate within the host cells. This also controls the copy number of linked DNA.
So, for getting many copies of the target DNA, it should be cloned in a vector whose origin support high copy number.
b. Selectable marker (marker gene)
– It is a gene that helps to select the transformants and eliminate the non-transformants.
– If a piece of DNA is introduced in a host bacterium, it is called transformation. Such bacterium is transformant. If transformation does not take place, it is non-transformant.
– Selectable markers of E. coli include the genes encoding resistance to antibiotics like ampicillin, chloramphenicol, tetracycline, kanamycin etc. Normal E. coli cells have no resistance against these antibiotics.
c. Cloning sites
– These are the recognition sites for restriction enzymes.
– To link the alien DNA, the vector needs a single or very few recognition sites.
– More than one recognition sites generate several fragments. It complicates the gene cloning.
– Ligation of alien DNA is carried out at a restriction site present in one of the two antibiotic resistance genes.
E.g. In vector pBR322, foreign DNA is ligated at Bam H I site of tetracycline resistance gene. As a result,recombinant plasmid is formed. If ligation does not occur, it is called non-recombinant plasmid.

– When a foreign DNA is inserted within a gene of bacteria,that gene is inactivated. It is called insertional inactivation. Here, the recombinant plasmids lose tetracycline resistance due to insertion of foreign DNA.
– When the plasmids are introduced into E. coli cells, 3 types of cells are obtained:
o Non-transformants : They have no plasmid. So they are not resistant to either tetracycline or ampicillin.
o Transformants with non-recombinant plasmid :
They are resistant to both tetracycline & ampicillin.
o Transformants with recombinant plasmid : They are resistant only to ampicillin.
– Recombinant plasmids can be selected out from nonrecombinant ones by plating transformants on ampicillin medium. Then the transformants are transferred on tetracycline medium.
– The recombinants grow in ampicillin medium but not on tetracycline medium. But, non-recombinants grow on the medium containing both the antibiotics.
– Thus, one antibiotic resistance gene helps to select the transformants. The inactivated antibiotic resistance gene helps to select recombinants.
– But this type of selection of recombinants is a difficult procedure because it needs simultaneous plating on 2 plates having different antibiotics. So, alternative selectable markers have developed based on their ability to produce colour in presence of a chromogenic substrate.
– In this, a recombinant DNA is inserted into the coding sequence (gene) of an enzyme, b-galactosidase. So, the gene is inactivated (insertional inactivation). Such colonies do not produce any colour. These are identified as recombinant colonies.
– If the plasmid in bacteria have no an insert, it gives blue coloured colonies in presence of chromogenic substrate.
d. Vectors for cloning genes in plants & animals Genetic tools of some pathogens can be transformed into useful vectors for delivering genes to plants & animals. E.g.
• Agrobacterium tumefaciens (a pathogen of many dicot plants) can deliver a piece of DNA (T-DNA) to transform normal plant cells into a tumor. These tumor cells produce the chemicals required by the pathogen.
The tumor inducing (Ti) plasmid of A. tumefaciens is modified into a cloning vector which is not pathogenic but can use mechanisms to deliver genes of interest into plants.
• Retroviruses in animals can transform normal cells into cancerous cells. So, they are used to deliver desirable genes into animal cells.
3. Competent Host (For Transformation with Recombinant DNA)
– Since DNA is a hydrophilic molecule, it cannot pass through cell membranes. So bacterial cells are made ‘competent’ to take up alien DNA or plasmid as follows:
– Treat bacterial cells with a specific concentration of a divalent cation (e.g. calcium) → DNA enters the bacterium through pores in cell wall → Incubate the cells with
recombinant DNA on ice → Place them briefly at 420C (heat shock) → Put them back on ice → Bacteria take up recombinant DNA.
Other methods to introduce alien DNA into host cells
• Micro-injection: In this, recombinant DNA is directly injected into the nucleus of an animal cell.
• Biolistics (gene gun): In this, cells are bombarded with high velocity micro-particles of gold or tungsten coated with DNA. This method is suitable for plants.
• ‘Disarmed pathogen’ vectors: They infect the cell and transfer the recombinant DNA into the host. E.g. A. tumefaciens.
PROCESSES OF RECOMBINANT DNA TECHNOLOGY
1. Isolation of the Genetic Material (DNA)
– Treat the bacterial cells/plant or animal tissue with enzymes like lysozyme (bacteria), cellulase (plants), chitinase (fungus) etc. The cell is broken releasing DNA & other macromolecules (RNA, proteins, polysaccharides & lipids).
– RNA is removed by treating with ribonuclease. Proteins are removed by treatment with protease. Other molecules are removed by appropriate treatments.
– When chilled ethanol is added, purified DNA precipitates out as a collection of fine threads in the suspension.
2. Cutting of DNA at Specific Locations
– Purified DNA is incubated with the restriction enzyme.
As a result, DNA digests. These DNA fragments are separated by a technique called gel electrophoresis.
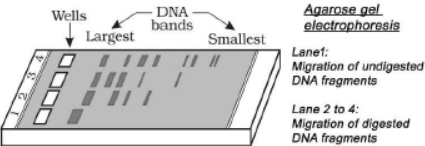
– Agarose gel electrophoresis is employed to check the progression of a restriction enzyme digestion. DNA is negatively charged. So it moves towards the anode. DNA fragments are separated according to their size through sieving effect of the agarose gel (a polymer extracted from sea weeds). The smaller sized fragment moves farther.
– The process is repeated with the vector DNA also.
– DNA fragments can be seen as bright orange coloured bands when they are stained with ethidium bromide and exposed to UV radiation.
– DNA bands are cut out from agarose gel. It is called elution. The cut-out gene of interest and cut vector are mixed and ligase is added. It creates recombinant DNA.
3. Amplification of Gene of Interest using PCR
– Polymerase Chain Reaction (PCR) is the synthesis of multiple copies of the gene of interest in vitro using 2 sets of primers & the enzyme DNA polymerase.
– Primers are small chemically synthesized oligonucleotides that are complementary to the regions of DNA.
Steps of PCR:
• Denaturation: It is the heating of target DNA (gene of interest) at high temperature (940 C) to separate the strands.
Each strands act as template for DNA synthesis.
• Annealing: It is the joining of the two primers (at 520 C) at the 3’ end of the DNA templates.
• Extension: It is the addition of nucleotides to the primer using a thermostable DNA polymerase called Taq polymerase. It is isolated from a bacterium, Thermus
aquaticus. It remains active in high temperature during the denaturation of double stranded DNA.
Through continuous replication, the DNA segment is amplified up to 1 billion copies.
The amplified fragment can be used to ligate with a vector for further cloning.

4. Insertion of Recombinant DNA into Host Cell
– Using any methods, the ligated DNA is introduced into recipient (host) cell / organism. They take up DNA from its surrounding.
– If a recombinant DNA bearing ampicillin resistant gene is transferred into E. coli cells, the host cells become ampicillin-resistant cells.
– If the transformed cells are spread on agar plates containing ampicillin, only transformants will grow. Untransformed recipient cells will die.
5. Obtaining the Foreign Gene Product
– The aim of recombinant DNA technology is to produce a desirable protein.
– If a protein encoding foreign gene is expressed in a heterologous host, it is called a recombinant protein.
– The cells with foreign genes can be grown in laboratory.
The cultures are used to extract the desired protein and purify it by using separation techniques.
– The cells can also be multiplied in a continuous culture system. Here, the used medium is drained out from one side while fresh medium is added from the other. It maintains the cells more physiologically active and so produces a larger biomass. It yields more desired protein.
Bioreactors
– These are the vessels in which raw materials are biologically converted to specific products, enzymes etc.,using microbial, plant, animal or human cells.
– Bioreactors are used to produce large quantities of products. They can process 100-1000 litres of culture.
– A bioreactor provides the optimal growth conditions (pH, temperature, substrate, salts, vitamins, oxygen) to get desired product.
– The most commonly used bioreactors are of stirring type (stirred-tank bioreactor). (Image 4)
It is usually cylindrical or with a curved base to facilitate the mixing of the reactor contents. The stirrer facilitates even mixing and oxygen availability. Alternatively, air can be bubbled through the reactor.
The bioreactor has
• An agitator system
• An oxygen delivery system
• A foam control system
• A temperature control system
• pH control system
• Sampling ports (for periodic withdrawal of the culture).
6. Downstream Processing
– It is a series of processes such as separation and purification of products after the biosynthetic stage.
– The product is formulated with suitable preservatives.
Such formulation undergoes thorough clinical trials and strict quality control testing.
